12
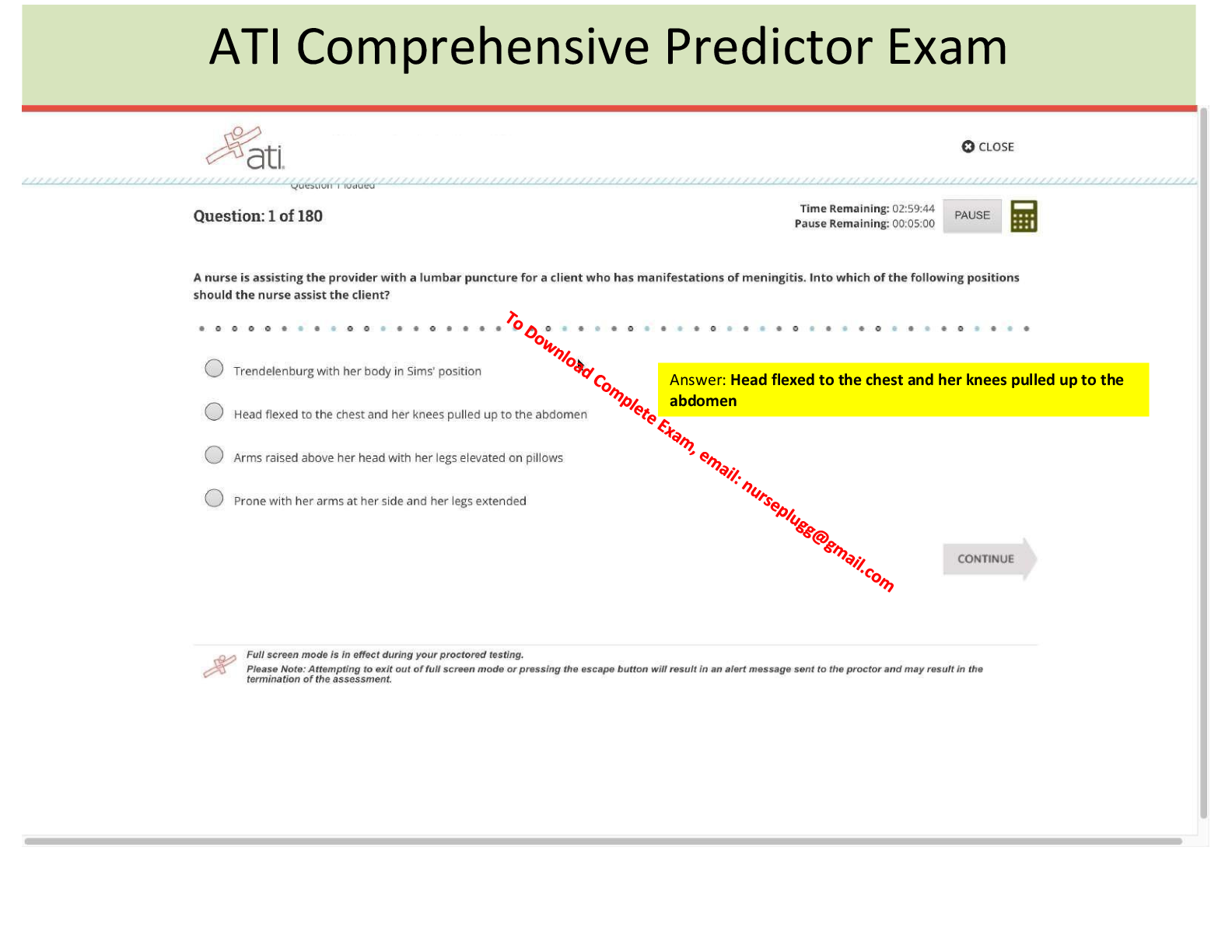
G1 to S to G2 to M
S is the synthesis
In anaphase the proteins that connect the sister chromatids
non chromosome kineticore elongate the cell
cleavage furrow is in animals
cell plate in plants (from golgi)
bact
...
12
G1 to S to G2 to M
S is the synthesis
In anaphase the proteins that connect the sister chromatids
non chromosome kineticore elongate the cell
cleavage furrow is in animals
cell plate in plants (from golgi)
bacteria undergo binary fission
dinoflag – envelope intact
diatoms – spindles within cell
S + G1 = SS
G1 + S = MM
G0 phase is inactive to continuing
Cyclin controls this cycle
growth factor – protein released that gets other cells to divide too
metastasis – spread of cancer cells
13
chiasmata – placing where crossing over occurred
14
pleiotropy – multiple effects of one gene
epistasis – a gene at one place alters the expression of another gene
polygenic inheritance – gradiant of traits
norm of reaction – phenotypic range
15
linked genes – tend to be inherited together
1 map unit is 1% recombinants max of 50%
nondisjunction – chromosomes don’t separate correctly
Types of chromosome problems – duplication, inversion, translocation, deletion
genomic imprinting – genes depend on mom or dad origin, caused by methylation
16
dead harmful DNA and alive harmless will kill
Semiconservative model is correct
Replication of bacterial DNAstarts at 1 point in both directions
makes bubbles with replication forks that fuse
uses DNA polymerase
DNA pol III goes from 5 to 3 of new strand
DNA ligase – joins okazaki
DNA pol I – replaces RNA primer with DNA
helicase – untwists double helix
topoisomerase – relieve strain ahead of fork’
single strand binding protein – bind to the unpaired DNA
primase – makes the RNA primer
DNA moves, not the enzymes
mismatch repair – cells use special enzymes to fix incorrect pairs
nuclease – cuts out bad part
This study source was downloaded by 100000790435732 from CourseHero.com on 10-05-2021 04:44:41 GMT -05:00
https://www.coursehero.com/file/478/Bio-FinalExamReview/
This study resource was
shared via CourseHero.com
telomeres – shitty genes at the end of DNA, telomerase catalyzes and restores
17
transcription – synthesis of RNA
Pre-mRNA to mRNA
Codons are writtin in the 5 to 3 direction
RNA polymerase goes from 5 to 3 (new strand)
promoter – starts here
terminator –ends here, polyadenylation sequence – cuts off mrna
transcription factors – mediate binding, help transcription
TATA Box – before the real sequence
pre to real mrna
5 cap – and a poly a tail – both facilitate export and help get rid of it
RNA splicing – gets rid of introns, spliceosome, snRNP’s recognize splice sites
Ribozymes – rna molecules functioning as enzymes
translation – synthesis of a polypeptide at ribosomes
anticodons, wobble (3rd base pair)
Ribosomes (with ribosomal RNA)
P to A to E
Small subunit finds start codon, gets large
Ribosomes go from 5 to 3 on the mrna
SRP – signal receptor protein bring ribosomes to ER
siRNA an mrRNA – regulate which genes are expressed
Mutations – silent , missense (still code but not for anything), nonsense (make a stop
codon)
19
histones – DNA packing
nucleosome – many histones in a beach
histone acetylation – helps code that gene more
methylation –deactivates
epigenetic inheritance – stuff like methylation that gets inherited
transcription factors – distal (enhances and use activators) and proximal
mrna degradation- dicers after miRNA
Protien degradation- tagged with ubiquitin and then proteasomes degrade them
21
totipotent – dedifferentiate and give rise to a different cell type
pluripotent – give rise to many but not all cell types
inducers – signal what the other cells should be too
egg polarity genes – control axis formation (along with morphogens)
segmentation groups – create segments
homeotic genes – appendages
apoptosis – programmed cell death
22
Linnaeus – taxonomy
Lamarck – evolution
homology – similarty due to common ancestry
23
This study source was downloaded by 100000790435732 from CourseHero.com on 10-05-2021 04:44:41 GMT -05:00
https://www.coursehero.com/file/478/Bio-FinalExamReview/
This study resource was
shared via CourseHero.com
Hard Wienberg – 2 alleles p and q create p2
+ 2pq + q2
= 1
Condition – large, no gene flow, no mutation, random mating
Genetic drift – populations are finite in size (bottleneck and founder)
Gene Flow – movement of fertile individuals, reduce differences in populations
frequency dependent selection – fitness declines when too common
24
anagenesis – stays 1
cladogenius –becomes 2
allopatric – geographically isolated
sympatric – no isolation geographically
heterochrony – change in development timing
allometric growth – relative rates of growth of different parts of the body
paedomorphosis – retain body features of juvenile
25
monophyletic – perfect
paraphyletic – not all decendents
polyphyletic – not all ancestors
shared primitive character – shared beyond a taxa
shared derived character – unique to a part
orthologous genes – passed in a straight line genes
paralogous – gene duplication, more than one copy
Archaen – Proterozoic – Paleozoic (first part is Cambrian) – Mesozoic- Cenozoic
29
Charophyceans – algeas and stuff before the bryophytes
cellulose, flaggelated sperm, sporopollenin
Bryophytes – seta, foot, capsule
micropyle – comes down stype and realeases two sperm
31
coenocytic – fungi that lack septa (walls dviding cells – porous)
haustoria – special hyphae that penetrate tissue of host
mycorrhizae – fungi that help plants
plasmogamy to a heterokaryon to a karyogamy
Chytrids – zoospores
Zygomycetes – zygosp
[Show More]





-2021 (1).png)





.png)






.png)